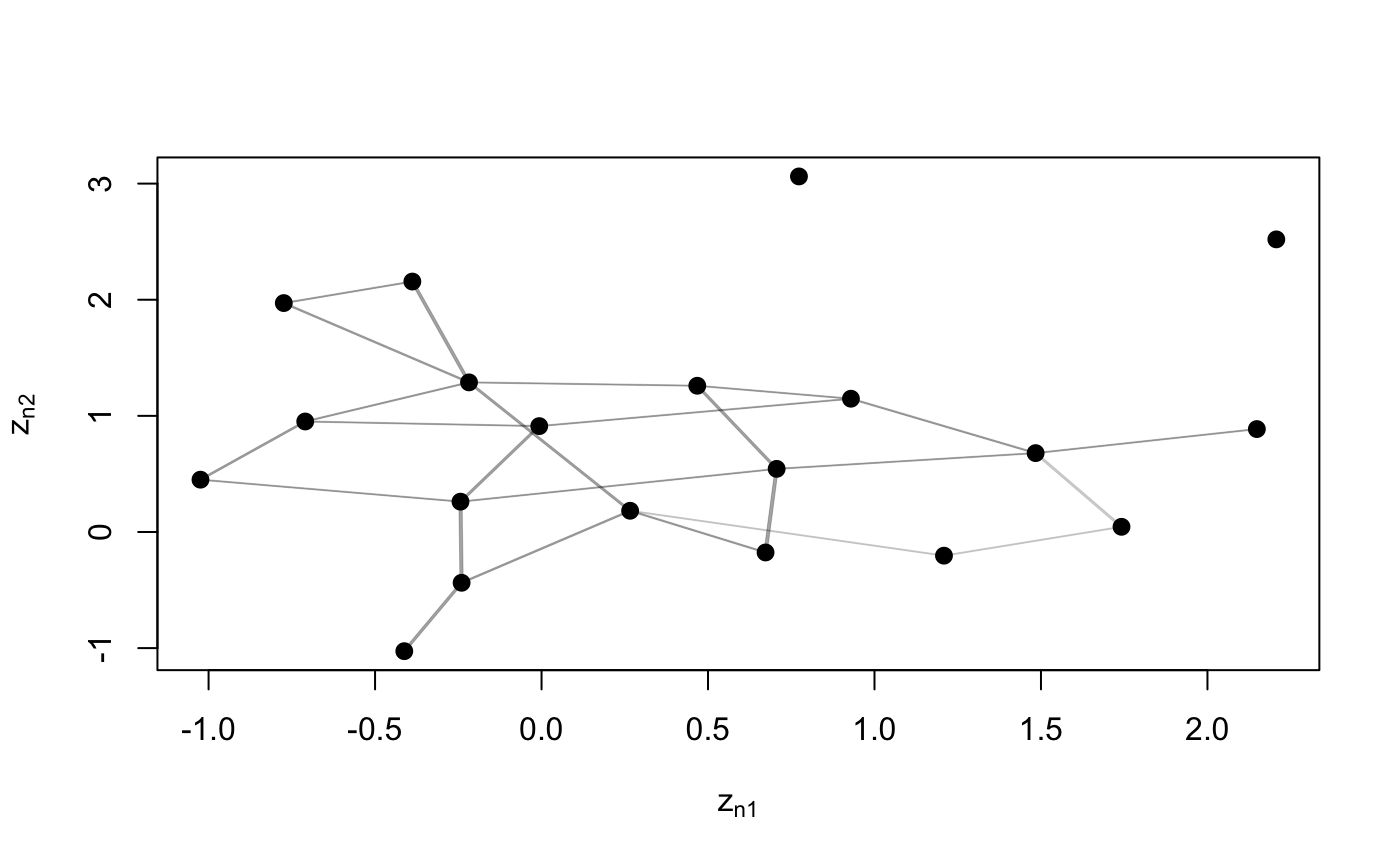
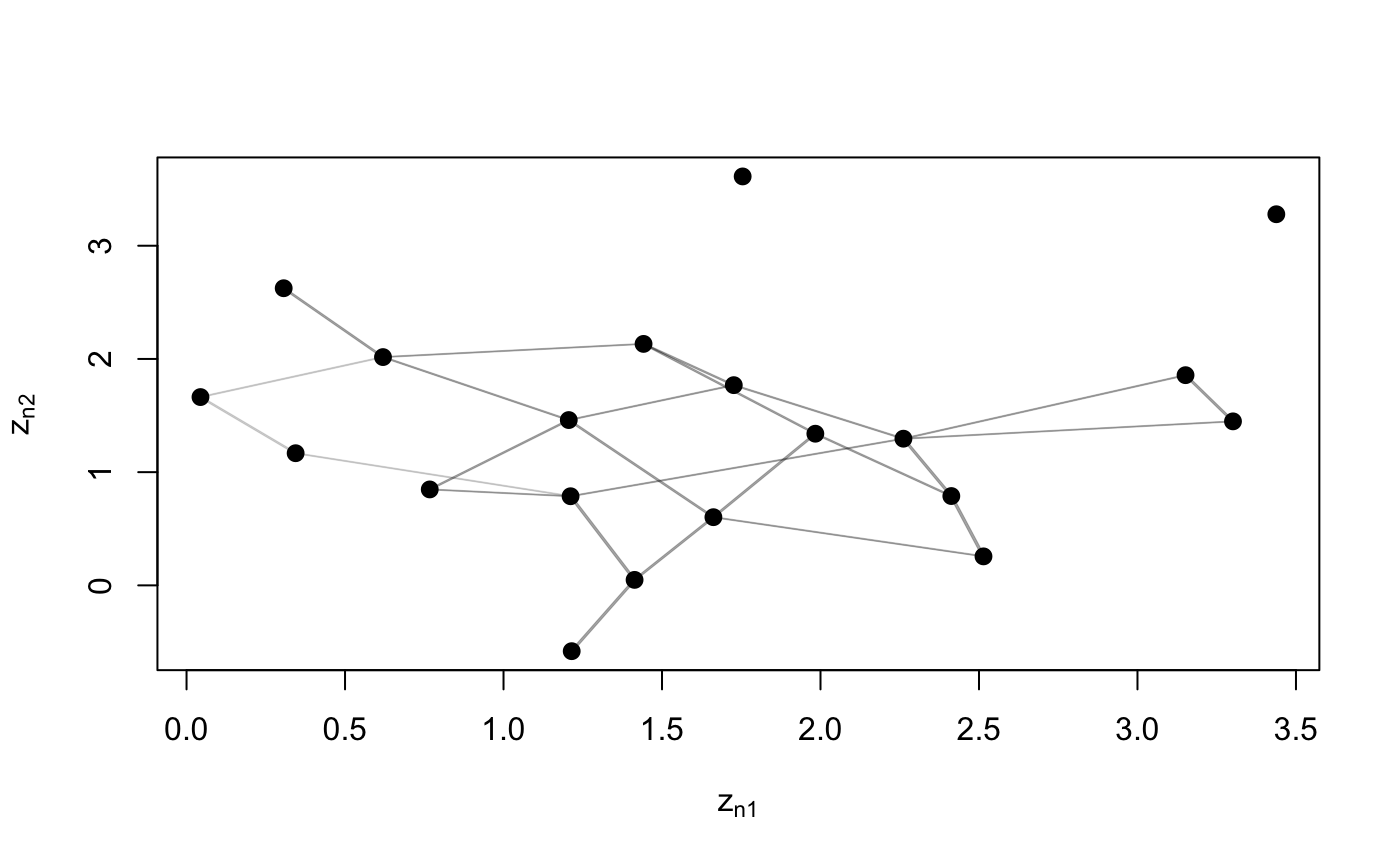
Function to plot the adjacency matrix of the network.
plotY(Y, Ndata = NULL, EZ = NULL, VZ = NULL, dimZ = c(1, 2), labels = NULL, colPl = 1, colEll = rgb(0.6, 0.6, 0.6, alpha = 0.1), LEVEL = 0.95, pchplot = 20, pchEll = 19, pchPl = 19, cexPl = 1.1, arrowhead = FALSE, curve = NULL, lwdLine = 0.3, xlim = NULL, ylim = NULL, verbose = FALSE, ...)
Arguments
| Y | list, or matrix containing a ( |
|---|---|
| Ndata | number of network views |
| EZ | posterior mean latent positions |
| VZ | posterior variance latent positions, if specified draw ellipse |
| dimZ | dimensions of Z to be plotted, default |
| labels | text to be added in the plot representing the labels of each node. Default |
| colPl |
|
| colEll |
|
| LEVEL | levels of confidence bounds shown when plotting the ellipses. Default |
| pchplot | Default |
| pchEll |
|
| pchPl |
|
| cexPl |
|
| arrowhead | logical, if the arrowed are to be plotted. Default |
| curve | curvature of edges. Default |
| lwdLine | lwd of edges. Default |
| xlim | range for x |
| ylim | range for y |
| verbose | if |
| ... | Arguments to be passed to methods, such as graphical parameters (see |
Examples
N <- 20 Y <- network(N, directed = FALSE)[,] plotY(Y)# Store the positions of nodes used to plot Y, in order to redraw the plot using # the same positions z <- plotY(Y, verbose = TRUE)plotY(Y, EZ = z)